Codility
Codility GenomicRangeQuery Solution (Python) Explained with Illustration
GenomicRangeQuery Solution (Python) Explained with Illustration
The prefix sum is a powerful concept to store the history statistics of an array by iterating the array once, so that later you can query the statistics of certain intervals of the array efficiently without iterating for each interval again.
GenomicRangeQuery test from Codility practice prefix sum in more intricate way, which is not that straight-forward to solve. Here I provide my solution with 100% correctness, the explaination will follow.
def prefix_sum(S):
index_map = {"A": 0, "C":1, "G":2, "T":3}
occurance_prefix_sum = [[0]*4 for _ in range(len(S)+1)]
for i, s in enumerate(S):
occurance_prefix_sum[i+1] = occurance_prefix_sum[i].copy()
occurance_prefix_sum[i+1][index_map[s]] +=1
return occurance_prefix_sum
def list_sub(l1, l2):
r = []
for a, b in zip(l1, l2):
r.append(a-b)
return r
def min_geno_factor(occurance):
geno_map = {0: "A", 1: "C", 2: "G", 3: "T"}
d = {"A": 1, "C":2, "G":3, "T": 4}
for i, count in enumerate(occurance):
if count != 0:
return d[geno_map[i]]
def solution(S, P, Q):
prefix_s = prefix_sum(S)
result = []
for p, q in zip(P, Q):
occurance = list_sub(prefix_s[q+1], prefix_s[p])
factor = min_geno_factor(occurance)
result.append(factor)
return result
Explaination
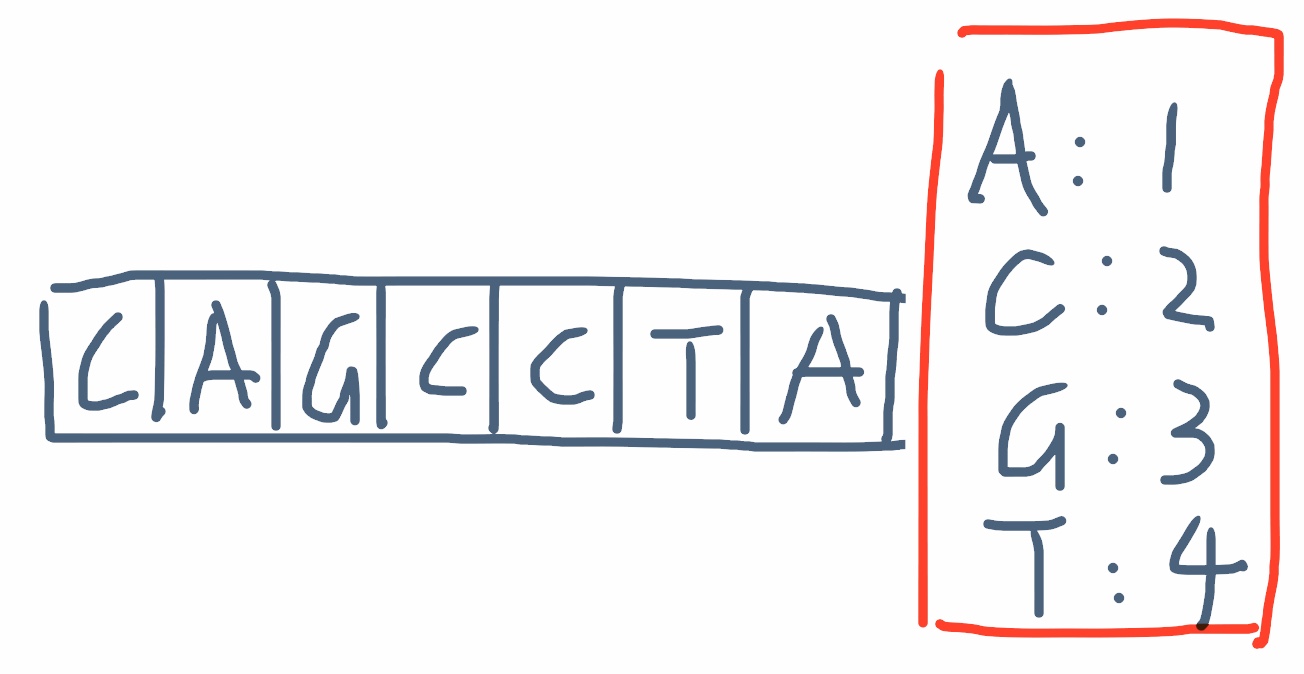
So let’s use the example case from Codility, [CAGCCTA] is our genomic sequence, with the factor {“A”: 1, “C”:2, “G”:3, “T”: 4}. The goal is to find the minimum factor for a given intervel. For example, if the interval is [2,4], then the selected geno is GCC and the one with lowest factor is C, so the solution should return 2.
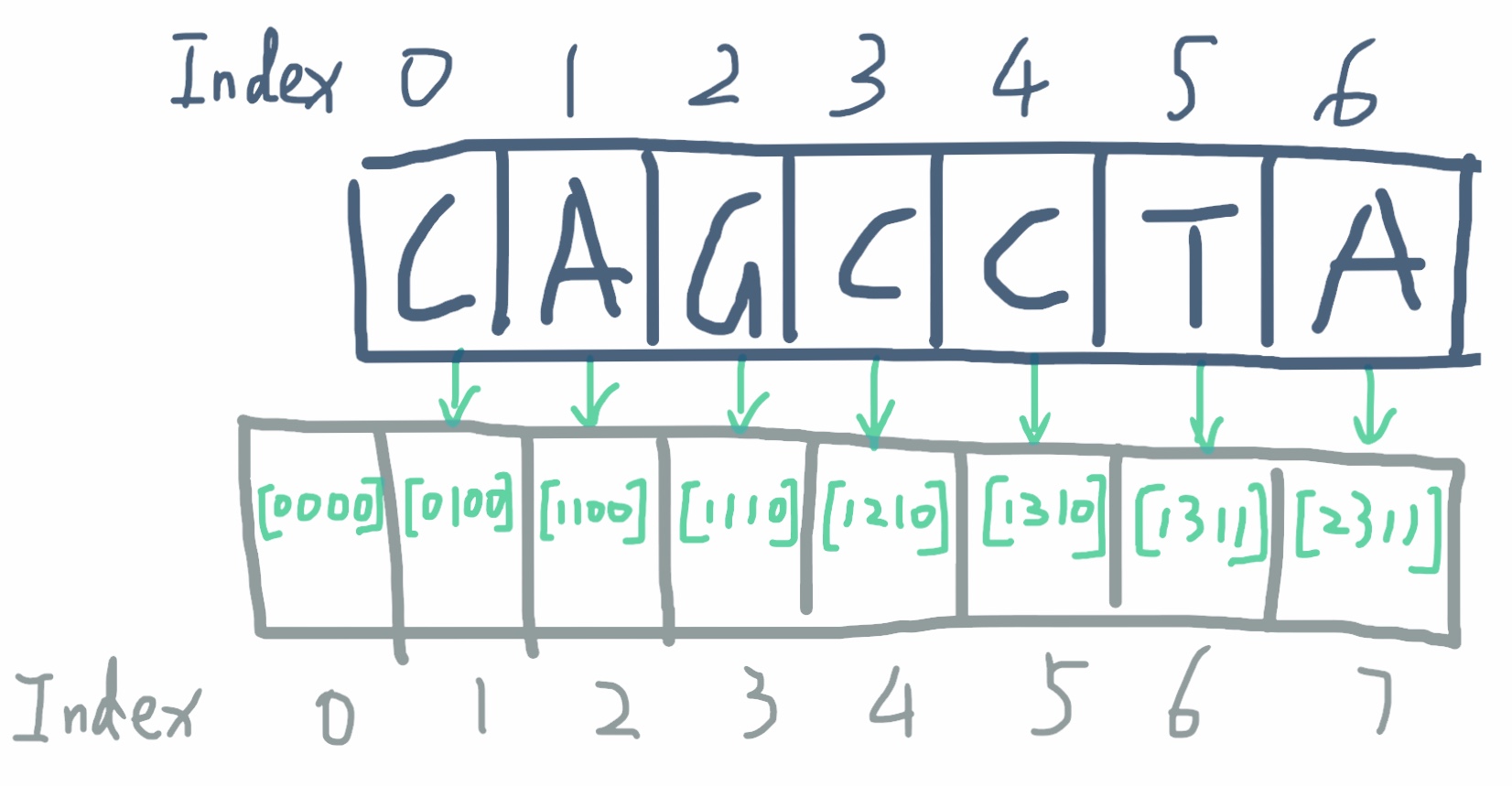
Unlike other prefix sum exercise, you can not sum up the charactors, and that has no meaning. Neither does it help to sum up over the factors, if we replace the geno with their factor respectively.
The trick is to sum up the number of occurance of each geno from left to right. So in the prefix sum array we do not store one value for each element but we store 4 values. In the end, we have an array of arrays. Each value in the 4-element array corresponds to the number of a geno seen so far, A occupy 0 position in the 4-element array, C occupy 1 position, so on so forth.
Note the prefix sum array has one more element than the original geno sequence, that is to store the [0,0,0,0] for easy substraction later.
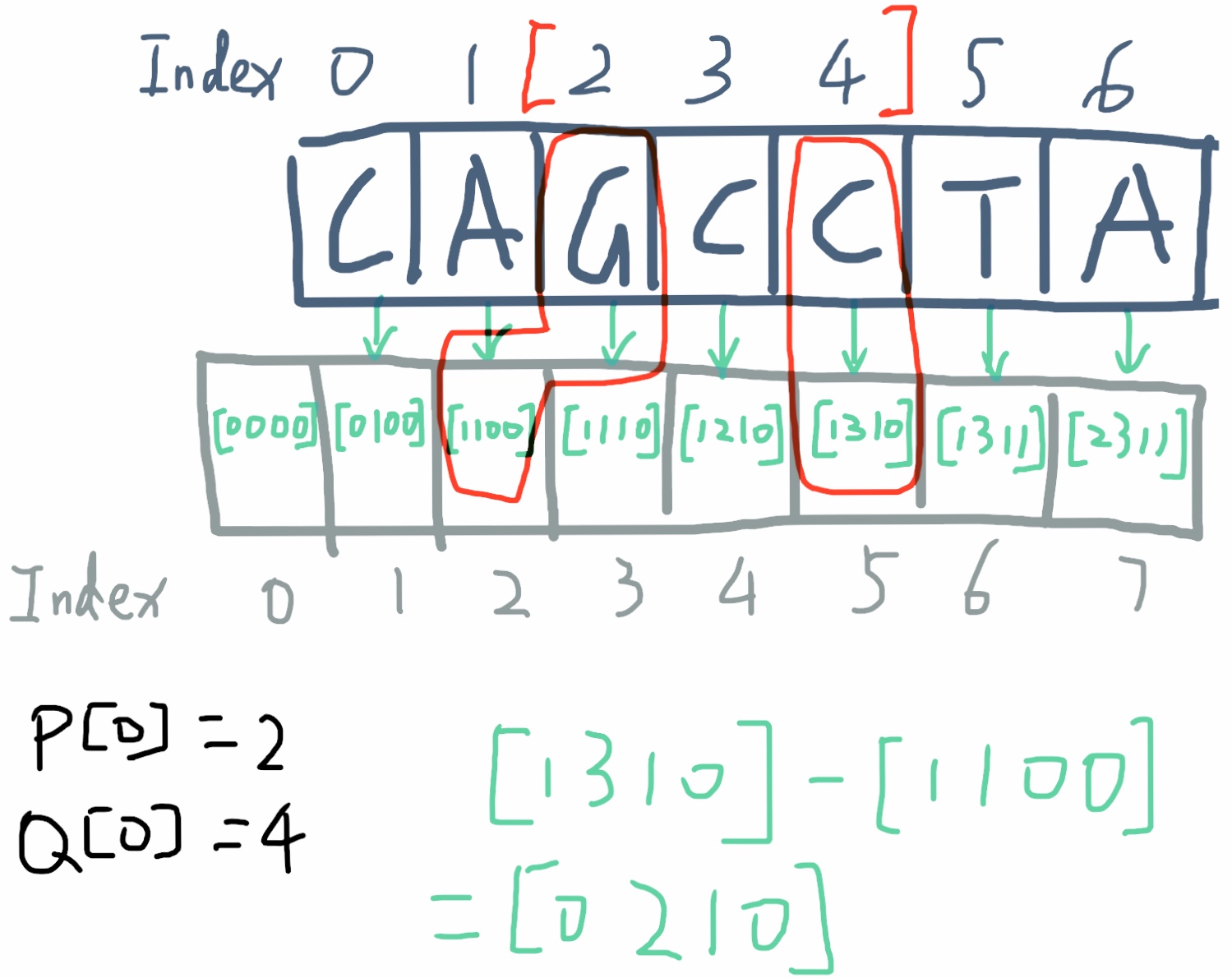
Given the prefix sum array containing the occurance count of each geno, it is now easy to calculate the minimum factor given an interval. For intervel [2,4], we take prefix_sum[4+1] - prefix_sum[2], and we can get the occurance count of the geno during this interval, [0,2,1,0]. C is one with the lowest factor 2 in the interval.
CODILITY · PYTHON